[BRH Data Resources] Help
![]() BRH Oda's Lab
BRH Oda's Lab
![]() BRHjapan(YouTube)
BRHjapan(YouTube)
[BRH Data Resources]
Help
Databases of Genome-based Research
Spiders/Butterflies/Insects/Arthropods/other animals
2025.6.12 NEW DATA from single-nuc RNAseq of late stage 5 & stage 7 have been added Click HERE.
2025.5.7 Single-cell/nucleus RNA-seq data for stage 7 embryos have been added in Pt_spider Genome Database v.3.
2023.5.20 A new function "Evolutionary characterization of Ptep genes has been added in Pt_spider Genome Database v.3.
2022.4.30 Single-cell/nucleus RNA-seq data have been added in Pt_spider Genome Database v.3.
2021.9.30 The Pt_spider Genome Database has been updated to version 3.
2021.7.20 Lecture slides used by Hiroki Oda in ISP2021 Osaka Univ are available from this link.
2021.4.28 A new BLAST search page was launched, whose databases consist of publicly available sequences from various spider species (see below).
2020.9.10 Our paper was published that included a set of genome-scale data obtained from hh, ptc and msx1 pRNAi embryos. These have been embeded in the genome browsers. Go to the article.
2020.9.10 "Library of data tables" page was created.
2020.3.16 A new function becomes available, which enables extraction of numeric data from the datasets deposited in GEO. Try an example to crick "Go to the datasets."
2019.8.30 RNAseq data on regions of the germ disc, which was obtained by Sawa Iwasaki-Yokozawa (unpublished), has been added in the Ptep1.0 genome browser. If interested to use it, contact us.
2019.8.14 The image/movie database updated. Click the words below! Try "time-lapse movies."
2018.11.5 A first version of our image/movie database is available.
2018.9.29 A link to "Insect Interaction Database" was added on this top page (see below).
2018.6.29 Iwasaki's developmental RNA-seq data were published.
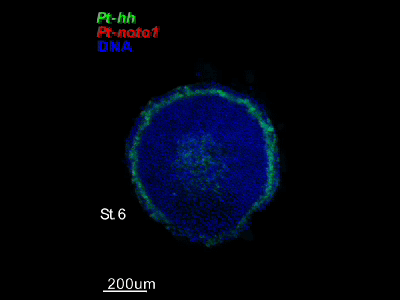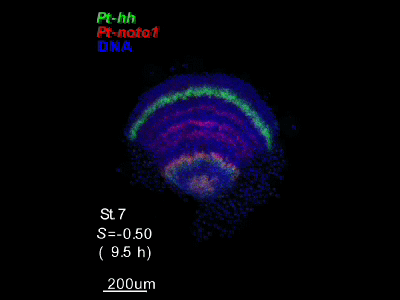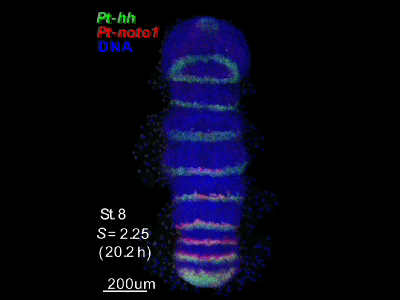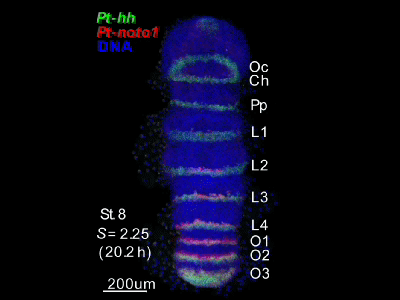
2018.4.16 An "Image Library" page has opened. Our hh-stripes paper was published.
2018.1.25 Ptep2.0 browser was launched with aug3 and NCBI Annotation Release 101 incorporated.
2017.11.28 Genome browser has been installed.
AUG3 trascript search has been linked to the genome browser.
2017.7.31 A paper on the genome of the common house spider Parasteatoda tepidariorum has been published.
2017.6.17 Our paper on the evolution of classical cadherin genes in arthropods has been published.
2017.6.8 The arthropod genome/RNA databases were updated.
 Adult female of the common house spider Parasteatoda tepidariorum
Adult female of the common house spider Parasteatoda tepidariorum